Level: ⚫⚫⚫⚪⚪ Advanced
Requirements
- Basic knowledge about git and GitHub
- A MAgPIE fork on GitHub
Content
- MAgPIE policy for updates of the main repository
- Procedure to create a MAgPIE pull request
Overview
The “pull request” – feeding back to the main repository
MAgPIE lives through its community of developers and is based on the idea that all research should be open, accessible and usable. While not enforced by any contract or license, we ask all developers to feed their model developments back to the main repository to make them more easily accessible to the whole community. While doing so means additional effort by the developer in order to fulfill the requirements for merging it into the main branch of the model (explained further down below), it comes with many advantages:
-
Developments get a higher visibility when put in the main repository compared to keeping them in a personal or even private repository. This increases the likelihood that the new research is taken up, further checked and finally cited in upcoming work.
-
Feeding your work back to the main repository will encourage others to do the same which will grant access to cool new features one otherwise would not get.
-
Feeding developments back ensures that others’ future developments are compatible with one’s own work, and avoids the situation where one’s own model becomes too difficult to merge into the main model version.
-
In publications you will be able to simply refer to a main release of the model rather than needing to explain how your specific version relates to the main line of the model.
Experience of the past years have shown us that:
- New features in the model will be taken up by researchers and leading to new and exciting research and,
- The researcher originally developing that feature will benefit from it through new collaborations and publications.
How do pull requests (PR) work?
Whenever you update your fork from the main repository (e.g. magpiemodel:develop) you use the
Git command pull. This attempts to merge the code from the main repository into your forked repository. A
“pull request” operates similarly, except from the other direction. By notifying
the maintainers of the main repository that you have code that you would like to merge, you start
a dialogue wherein the code can be reviewed and tested before it is incorporated into the main repository.
For MAgPIE, this process takes place over GitHub.com. After reviewing your code, the maintainers “pull”
from your repository into the main repository.
Steps to submitting a pull request
Below is a step-by-step guide to submitting a pull request, from committing to your own fork
to filling out the PR template itself. For the purposes of this tutorial, the PR includes a
new output script, FSEC_environmentalPollutants.R.
On your local fork
Complete and thoroughly test your code changes
When actively developing MAgPIE, you should refrain from making additions to the master branch.
In general one should either create branches off of your fork of develop branch for specific features, or
change the fork’s develop branch itself.
When developing the MAgPIE model, please following the coding etiquette available here.
This is the output script, currently on the develop branch of my local, forked repository
(mscrawford/magpiemodel:develop), that will be included into the magpiemodel:develop branch:
# | (C) 2008-2022 Potsdam Institute for Climate Impact Research (PIK)
# | authors, and contributors see CITATION.cff file. This file is part
# | of MAgPIE and licensed under AGPL-3.0-or-later. Under Section 7 of
# | AGPL-3.0, you are granted additional permissions described in the
# | MAgPIE License Exception, version 1.0 (see LICENSE file).
# | Contact: magpie@pik-potsdam.de
# --------------------------------------------------------------
# description: Create FSEC environmental pollutants output dataset
# comparison script: FALSE
# ---------------------------------------------------------------
# Version 1.00 - Michael Crawford
# 1.00: first working version
library(lucode2)
library(magpie4)
message("Starting FSEC environmental pollutants output runscript")
############################# BASIC CONFIGURATION #######################################
if (!exists("source_include")) {
title <- NULL
outputdir <- NULL
# Define arguments that can be read from command line
readArgs("outputdir", "title")
}
#########################################################################################
message("Script started for output directory: ", outputdir)
load(file.path(outputdir, "config.Rdata"))
title <- cfg$title
message("Generating environmental pollutants output for the run: ", title)
gdx <- file.path(outputdir, "fulldata.gdx")
baseDir <- getwd()
pollutantsOutputDir <- file.path(baseDir, "output", "pollutants")
if (!dir.exists(pollutantsOutputDir)) {
dir.create(pollutantsOutputDir)
}
out <- getReportGridPollutants(gdx = gdx, reportOutputDir = pollutantsOutputDir, magpieOutputDir = outputdir, scenario = title)
In particular, at this point you should pull the most recent magpiemodel:develop branch
into your repository. This ensures that your changes are already embedded into the current
magpiemodel:develop branch and will streamline the process of testing your innovations and
later merging them.
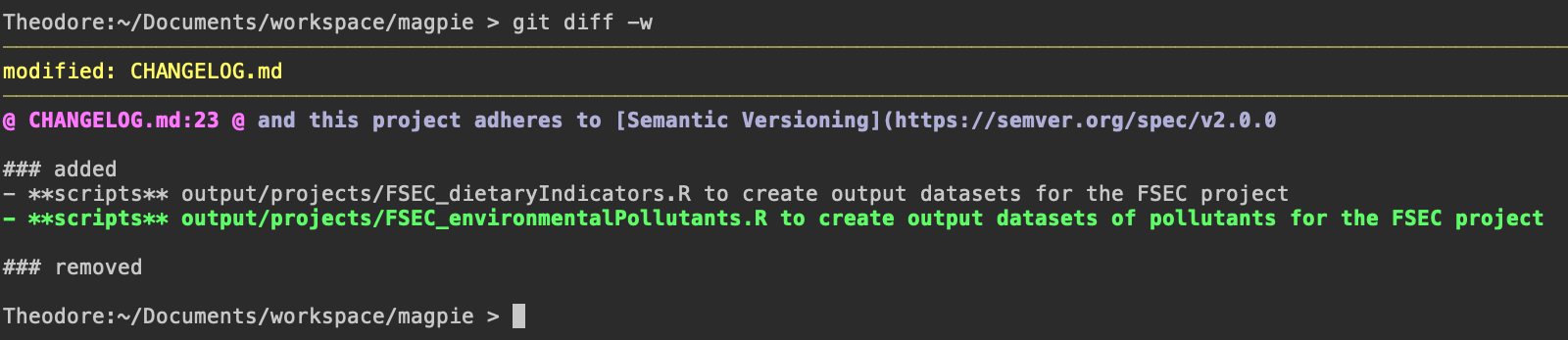
Modify the CHANGELOG.md
It’s critically important that you add your various changes to the CHANGELOG.md in the base
directory of your fork. This enables us to trace developments over time and, eventually,
package them cleanly into release candidates. Below is the current state of the unreleased MAgPIE
develop branch:
# Changelog
All notable changes to this project will be documented in this file.
The format is based on [Keep a Changelog](https:/keepachangelog.com/en/1.0.0/),
and this project adheres to [Semantic Versioning](https:/semver.org/spec/v2.0.0.html).
## [Unreleased]
### changed
- **scripts** added new disaggregation script to provide grid cell level BII
- **inputs** updated non-agricultural water use scenarios (watdem_nonagr_grper.cs3)
- **config** included switch for non-agricultural water demand (s42_watdem_nonagr_scenario) in scenario_config.csv
- **config** included SHAPE SDP scenarios in scenario_config.csv
- **34_urban** added set urban_scen34 and the switch c34_urban_scenario
- **35_natveg** corrected naming of Frontier Forests (FF) to Intact Forest Landscapes (IFL) and changed input data for BH_IFL implementation.
- **scripts** replaced redundant files config.log and config.Rdata with a config.yml
- **scripts** removed test script "irrig_dep_test" from "start" folder to "extra" folder
### added
- **scripts** output/projects/FSEC_dietaryIndicators.R to create output datasets for the FSEC project
- **scripts** output/projects/FSEC_environmentalPollutants.R to create output datasets of pollutants for the FSEC project
### removed
### fixed
- **inputs** included data for Sudan
- **18_residues** off realization; missing variable declarations
- **34_urban** exo_nov21 realization; bugfix in calculation of biodiversity value
- **50_nr_soil_budget** off realization; missing variable declarations
- **59_som** static realization; avoid division by zero
- **62_material** exo_flexreg_apr16 realization; avoid division by zero
- **80_optimization** nlp_par realization; bugfix i2 in submission loop
- **scripts** calibration; set NA values to 1
- **scripts** fixed misleading warning in check_config
Note that the CHANGELOG separates all code changes into changed, added, removed, and fixed. If you made multiple
commits which included changes, additions, and removals, these must be broken down here. In practice, it is
better git etiquette (i.e. nicer for the maintainers) to have smaller, discrete PRs that implement a singular feature or advancement, than larger PRs implementing many disconnected features simultaneously.
In particular I have added a single line, detailing the addition of the FSEC_environmentalPollutants.R
output script.
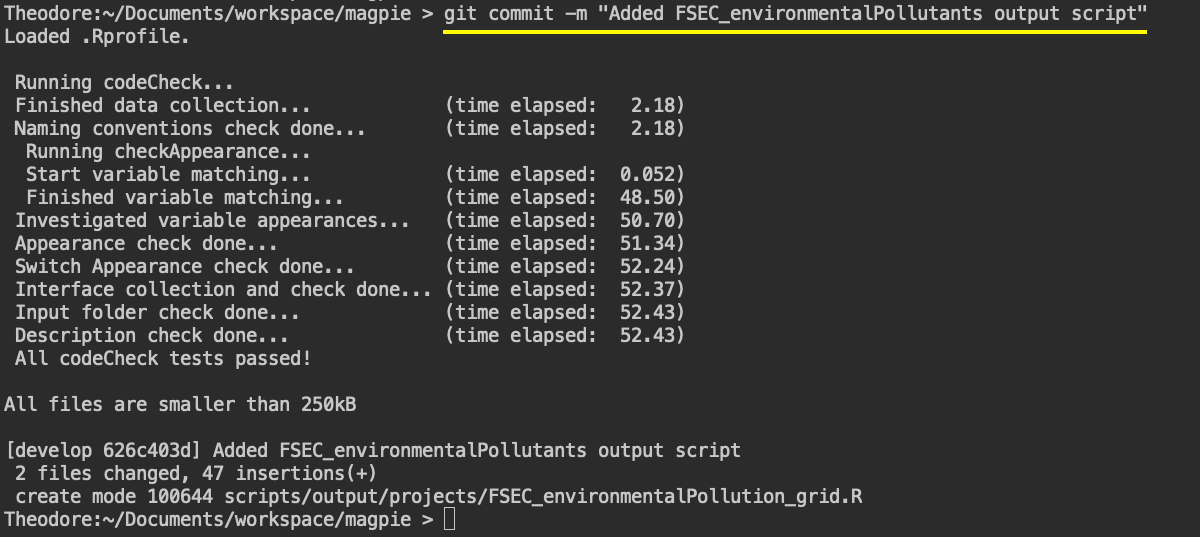
Commit your changes to your local fork
With my added output script and corresponding update to the CHANGELOG.md, I need to commit my changes and push
them to my own fork. I first staged the changes (by running git add * in the terminal) and now commit them:
Push the new commit to your repository on GitHub
Unless you’ve heavily customized your fork, you will want to push this new commit to origin:develop, as below:
On your GitHub account
Creating a new PR
From here we initiate the PR from github.com. Navigating to my own fork, we see my new commit is represented:
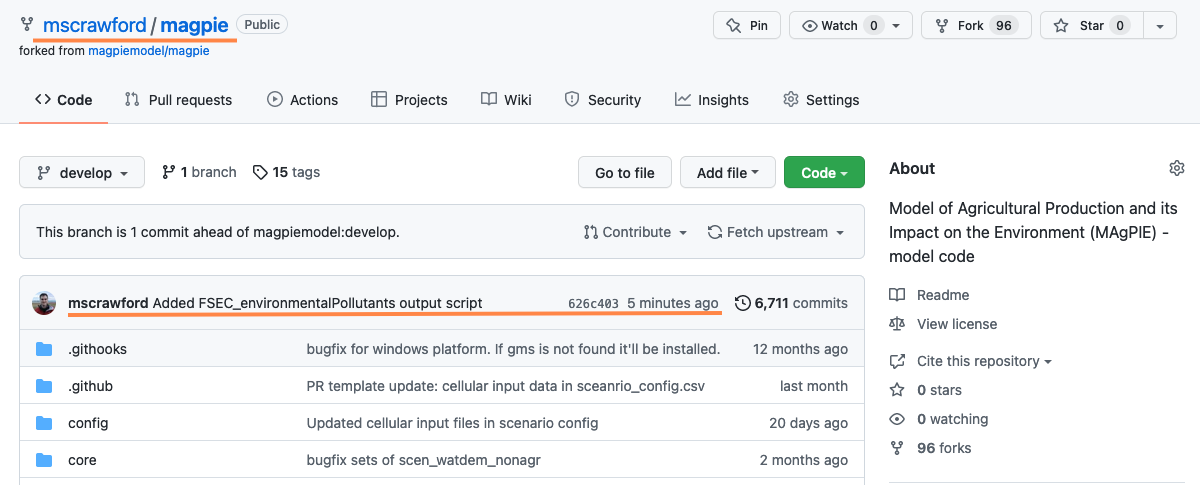
One can futher observe that my branch is 1 commit ahead of magpiemodel:develop.
I now navigate to Pull requests in the top-left menu, clicking New pull request.
Now is a key step wherein you should check a number of details before creating your new pull request.
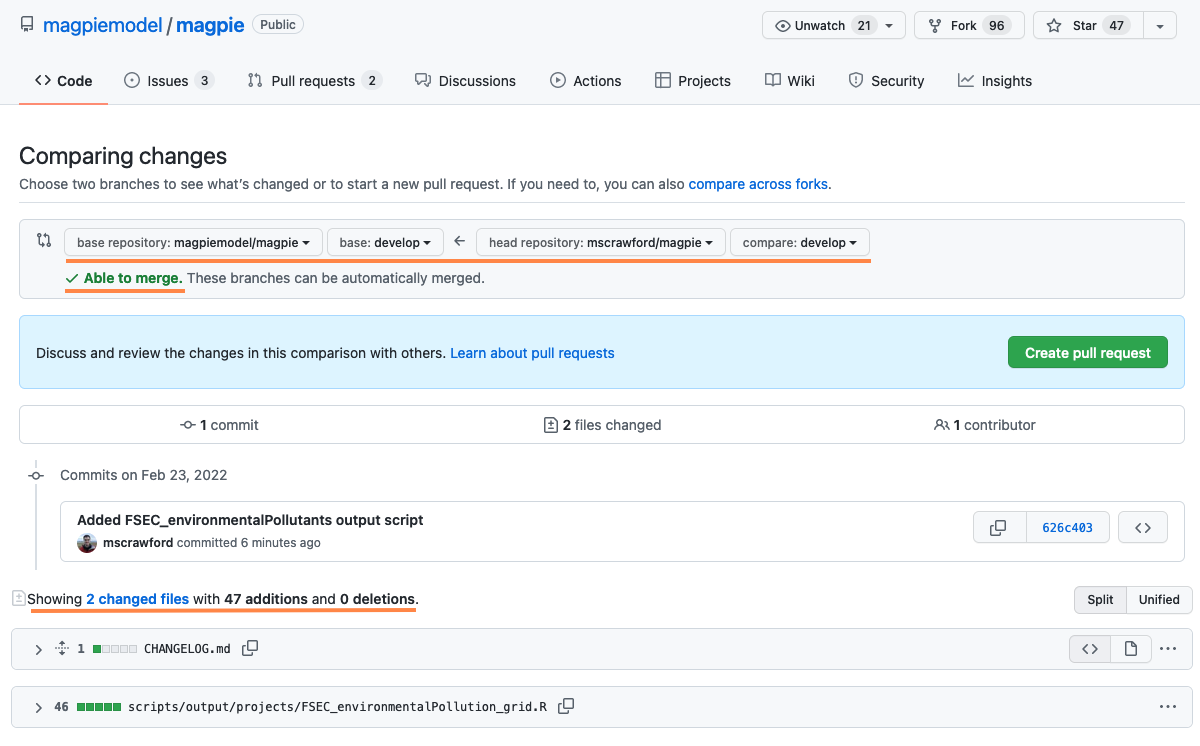
- Verify that you are merging from your correct, forked, branch to the correct branch of the
magpiemodel/magpierepository (this should almost without exception be thedevelopbranch). - Check that your changes are as expected, and fully represented. For example, if want to merge multiple commits, are they all there?
- Can the two branches be automatically merged? If not, you need to pull the most recent
magpiemodel:developbranch and merge these latest changes into your own branch.
A tip: It is possible to pull from the
magpiemodel:developbranch to your fork (likelyorigin:develop) through GitHub.com, and then you could pull from the origin to your local machine. Another potentially convenient method could be adding anupstreamremote the your git repository on your local machine, as here.
The PR template
The PR template is a space for you to describe the changes you’ve made to the MAgPIE model and provide information for MAgPIE’s maintainers to review the code and assess the level of detail needed to safely integrate it into the main repository. One should go through the template thoroughly and address each checkbox.
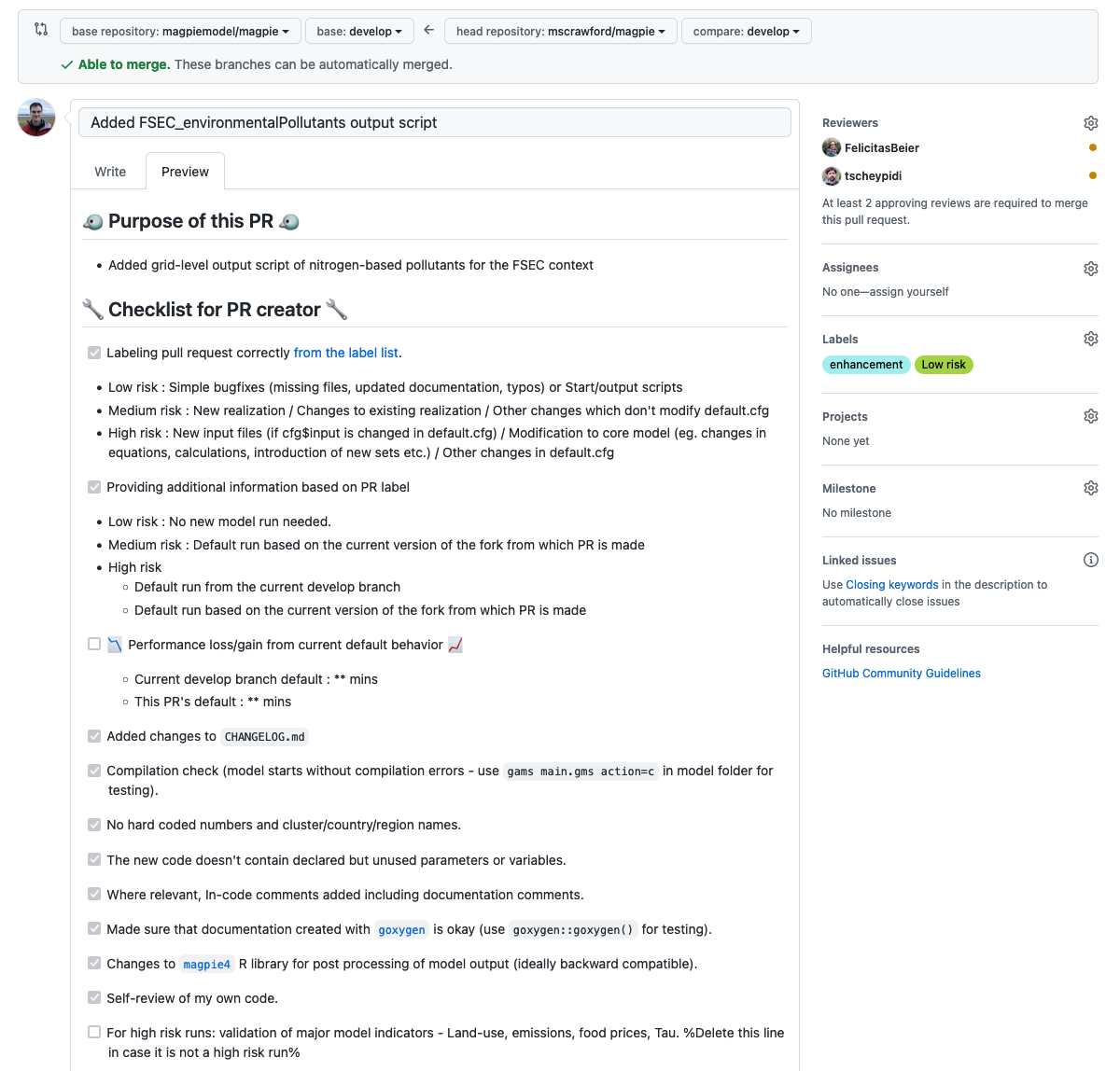
A breakdown of the PR template
- Label your pull request. The amount of time invested into validating your code changes scales with the
risk in merging them. In general, changing input files and the core model are very high risk changes and need special
attention. Shown in this tutorial however, adding start scripts are a low risk change. Therefore I add the
low risklabel from the label list (by clicking the wrench on on the right). - Provide additional information based on the PR label.
- Performance gain/loss. For medium and high risk changes, one must report the runtime of the new branch. High risk
in particular should be compared with a control run of the
magpiemodel:developbranch. Ideally additions to the code should not incur runtime changes. If they do, there must be good reasons for it. - Added changes to
CHANGELOG.md. - Compilation check. Make sure that your model compiles successfully. Navigate to your
magpiedirectory and within the console rungams main.gms action=c.
A tip: It may be necessary - if you haven’t already run the model in your directory - to download the input data from the PIK servers. To do so, run
Rscript start.Rand select3: download data | just download default.cfg input data.
- No hard-coded numbers and cluster/country/region names. MAgPIE is able to run across flexible cluster, country, and region definitions. This flexibility stems from these definitions only entering the model through input parameters as the model is being started. Thus, hard-coding these definitions will lead to erronous results.
- The new code doesn’t contain declared but unused parameters or variables.
- Where relevant, In-code comments added including documentation comments.
- Make sure that the documentation worked for your additions.
- Changes to the
magpie4enable postprocessing of any new model output.magpie4is the R repository which is used to generate analyses from the model output datasets. - Self-review of my own code. Perhaps the most important checkbox. Are you certain that your code does what you think it does?
- For high risk runs: validation of major model indicators - Land-use, emissions, food prices, Tau. Screenshots of
the Shinyapp at the with regional level for a default run of both your new developments and the current
magpiemodel:developbranch are appropriate for most changes.
After completing the PR template you should request at least two reviewers from the MAgPIE mainanence team.
The review process
After recieving your PR request, both MAgPIE maintainers will independently review your changes and request clarifications or changes as appropriate.
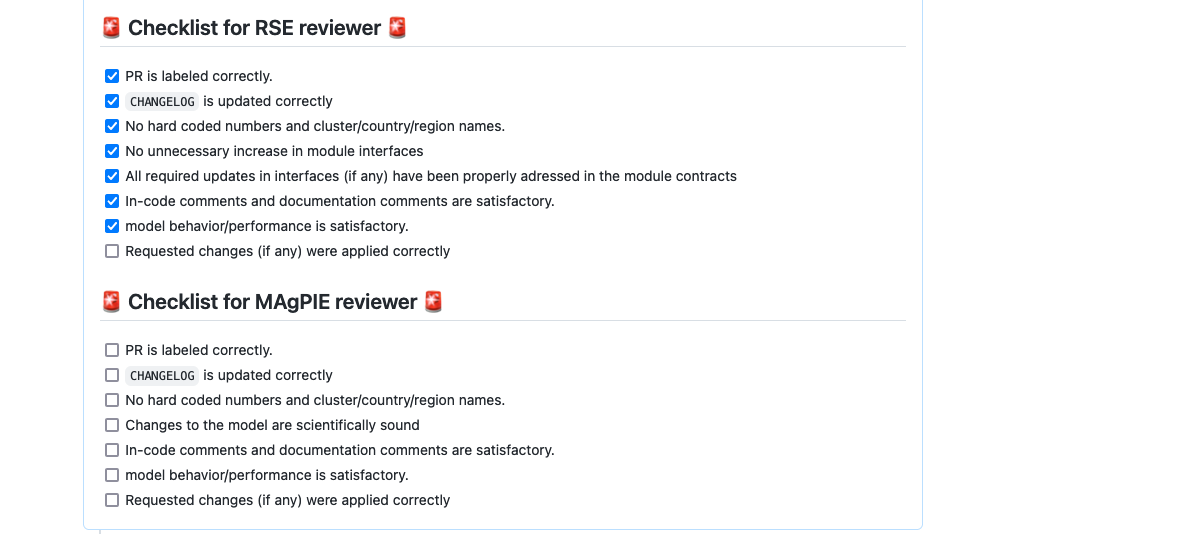
The reviewers, one of whom must be from the Research and Software Engineering (RSE) department at PIK, use the above
checklist. After this interaction has been completed, they will accept the PR and it will be automatically merged
into the magpiemodel:develop branch.
Exercises (click on the arrows to uncover the solution)
Produce figures depicting the current `magpiemodel:develop` branch's behaviour for these indicators: Land-use, emissions, food prices, Tau.
- Open a terminal in the main MAgPIE model folder
- Ensure that you are on the
developbranch by runninggit checkout develop- Run a default run with the start script (
Rscript start.R)- Within the main model directory, in an
Rsession runshinyresults::appResultsLocal()You may have to install this package on your local machine.- Print these following plots to .pdfs:
- Resources/Land Cover/+/Cropland
- Resources/Land Cover/+/Pastures and Rangelands
- Emissions/CO2/Land
- Productivity/Landuse Intensity Indicator Tau
- Prices/Food Price Index
Ensure that your GAMS code compiles.
- Open a terminal and navigate to your main MAgPIE directory
- Ensure that you have checked out the correct branch
- Run
Rscript start.Rand download the input data (3: download data)- Within your terminal, run
gams main.gms action=cEnsure that the documentation for MAgPIE is successful derived
- Open an R session within your model’s main directory
- Run
goxygen::goxygen()
Creating a new realization Overview Input data repositories & Patches
